Abstract
Background
The 5-year survival rate for acute myeloid leukemia (AML) remains poor with most patients succumbing to relapse or refractory disease. Recently, the BCL2 specific inhibitor venetoclax has shown promising anti-leukemia activity in high-risk AML patients. Most patients, however, ultimately develop resistance to monotherapy and novel combination treatments with venetoclax are needed for patients with no other therapy options available. In this study we identified high expression of calcium binding protein S100A8/S100A9 to be associated with venetoclax resistance and looked for drug combinations to overcome the resistance in AML patient samples overexpressing S100A8 and S100A9.
Methods
Gene expression was assessed by RNA sequencing of AML patient samples and validated by qPCR. Gene enrichment analysis was performed on differentially expressed genes between venetoclax highly sensitive (n=3) and resistant (n=4) samples. Sensitivity of AML patient derived mononuclear cells was assessed to 304 different small molecule inhibitors by measuring cell viability after 72 h incubation with 5 different concentrations (1-10,000 nM) using the CellTiter-Glo (CTG) assay. A drug sensitivity score (DSS) was calculated based on a modified area under the dose response curve calculation. Drug combination studies were performed using AML cell lines resistant to venetoclax and confirmed with primary patient cells (n=15). Data of the drug combination studies were analyzed with the Zero Interaction Potency (ZIP) model by considering a dose-response matrix where two drugs are tested at 8 concentrations in a serially diluted manner. Statistical dependence between gene expression and drug sensitivity data was assessed by Pearson's correlation coefficient modelling.
Results
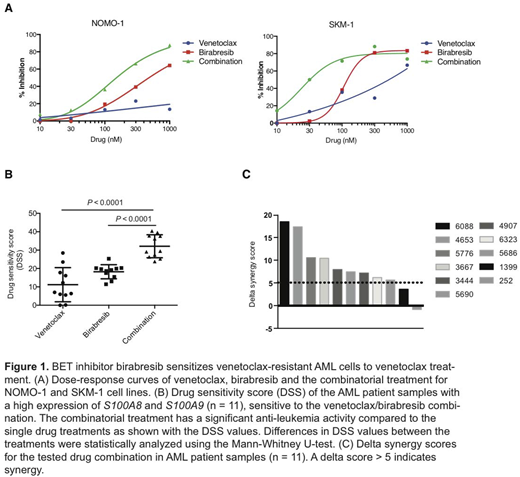
Venetoclax resistant AML patient samples were found to overexpress genes related to immune responses including inflammatory related proteins S100A8 and S100A9. The expression of S100A8 and S100A9 was upregulated in a sub-group of AML patients with somatic mutations in DNA methylation genes IDH2 and TET2 and chromatin modifier ASXL1. Functional studies with AML cell lines validated high expression of the S100 proteins in cells insensitive to venetoclax (NOMO-1, SKM-1 and SHI-1) whereas sensitive cell lines (MOLM-13, Kasumi-1 and ML-2) did not express the proteins. Integrated analysis of S100A8 and S100A9 expression and ex vivo drug sensitivity data indicated positive correlation of S100 expression with sensitivity to BET inhibitor (birabresib), PI3K inhibitor (TG100-115) and MEK1/2 inhibitor (AZD8330). In contrast, sensitivity to venetoclax and the FLT3 inhibitor quizartinib negatively correlated with S100 gene expression. Subsequently, we combined positively correlating drugs with venetoclax and tested the efficacy of these combinations in AML cell lines and patient samples. From the drug combination studies we found that BET inhibitor birabresib was able to overcome resistance to venetoclax treatment. The BCL2/BET inhibitor combination was synergistic in venetoclax resistant cell lines NOMO-1 and SKM-1, which express high-levels of S100A8 and S100A9 (Figure 1A). Efficacy of the combination on primary AML patient samples correlated with the expression level of the S100 genes. Nine of 11 high expression samples were sensitive to the venetoclax/birabresib combination (Figure 1B-C), whereas no synergy was observed in 3 of 4 samples with a low level of S100 expression.
Conclusions
The calcium binding proteins S100A8 and S100A9 are abundant in myeloid cells and are associated with poor prognosis in AML (Edgeworth et al, J Biol Chem. 1991; Nicolas et al, Leukemia 2011). From ex vivo and in vitro analyses of AML, we found that high expression of S100A8 and S100A9 is highly correlative with resistance to the BCL2 inhibitor venetoclax. In contrast, high S100A8 and S100A9 expression correlates with sensitivity to BET inhibitor birabresib. Interestingly, our studies showed that these two drugs act synergistically in venetoclax resistant AML cell lines and AML patient samples and may be a beneficial novel combination that should be further confirmed in preclinical and clinical investigations.
Porkka:Celgene: Honoraria, Research Funding; Novartis: Honoraria, Research Funding. Heckman:Orion Pharma: Research Funding; Novartis: Research Funding; Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal